LabKey Server is a Java-based web application. Recent changes to Oracle’s Java licensing model and release schedule require changes by software providers like LabKey, as well as application administrators. Below LabKey’s VP of Product Strategy, Adam Rauch, explains the recent changes, how LabKey plans to address them, and what actions need to be taken by teams running LabKey Server to ensure on-going stability and support.
Recent Changes to Java Release Cadence and Licensing
Last year, Oracle announced several significant changes to the Java release and support model that introduce complexity to the previously straightforward process of deploying a Java runtime. Organizations will need to make decisions and revise their upgrade processes, but we believe these changes will lead to a stronger Java platform, one that will be more responsive and easier to support in the long term.
Perhaps the biggest news is that Oracle now requires a paid subscription for all use of the Oracle Java Runtime (“Oracle Java SE”) in production environments. Starting with Java 11 (released September 2018), organizations are required to pay up to $25 per CPU per month for production server or cloud use of Oracle Java SE. This seems to apply to everyone… no exceptions for academic, non-profit, or small organizations. With a subscription, Oracle will provide long-term support (LTS) for designated versions of their runtime (Java SE 8, 11, 17).
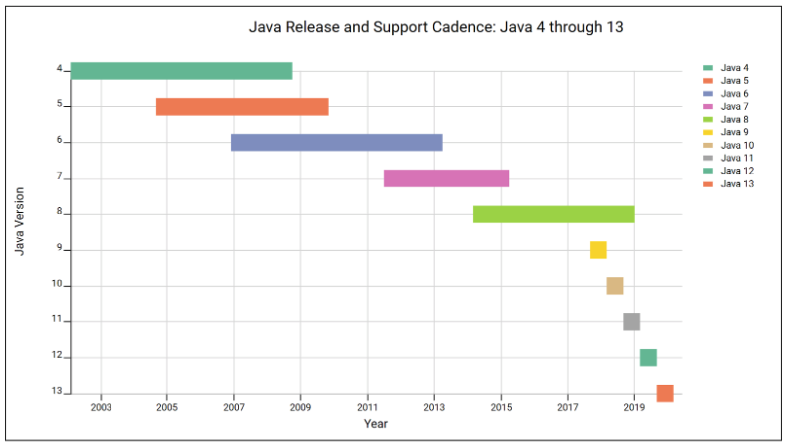
With Java 9, the release cadence transitioned from major versions every five or six years to more incremental feature releases every six months. According to Oracle, this new time-driven release model allows more rapid innovation, but it also means organizations will need to upgrade more quickly to keep pace with the changing platform.
This diagram (brought to you by the LabKey Visualization API) helps illustrate this change, with each bar capturing the time Oracle publicly supported (or plans to support) each version:
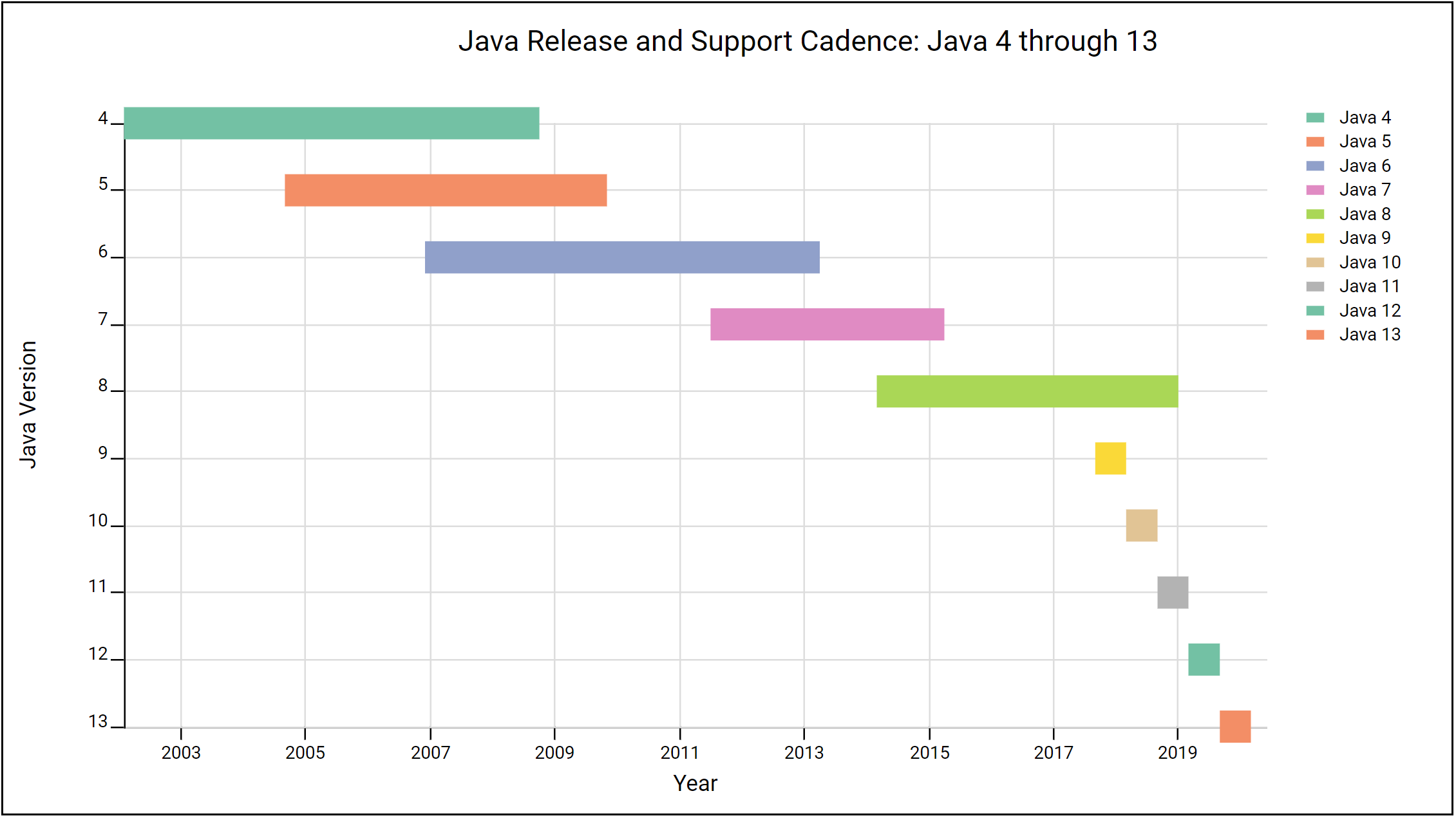
Note that in addition to more rapid and shorter releases, Oracle has eliminated the overlap between versions. Under the new model, public support for previous releases ends immediately upon release of a new version. As a result, Java 9 was end-of-lifed (no further support) the day Java 10 was released and Java 10 was end-of-lifed the day Java 11 was released.
Other Options and LabKey’s Response
As a Java-based web application, every deployment of LabKey Server is affected by these changes. You can certainly move to a paid subscription with Oracle, but many of you have told us you want a free Java option. LabKey has heard you and, starting with LabKey Server release 18.3, you now have the ability to deploy with a completely free, open-source Java runtime.
This is possible because Oracle has embraced OpenJDK, the open source implementation of the Java platform. Oracle recently contributed its remaining commercial features to the OpenJDK project and now builds its subscription Oracle Java SE from the OpenJDK sources. In fact, Oracle distributes two versions of OpenJDK: the Oracle Java SE requiring the commercial license and a production-ready open-source build of OpenJDK licensed under GPLv2 with the “Classpath Exception” (“Oracle OpenJDK”). The code is the same; the Oracle OpenJDK distribution merely lacks the long-term support provided under the subscription. Oracle might add proprietary enhancements (e.g., advanced garbage collection algorithms, just-in-time compilation, profilers, and other tools) to future commercial runtimes, but at the moment, these distributions should be interchangeable.
In response to these changes and our clients’ requests, LabKey has shifted our development, testing, and hosting to focus on Oracle OpenJDK 11. We will continue limited testing on the commercial Oracle Java SE releases, but the vast majority of our attention will be focused on OpenJDK. We plan to support future OpenJDK releases at or shortly after they’re released. Where possible, we’ll hotfix the current production release of LabKey Server to ensure it’s compatible with newly released versions of OpenJDK. (For example, even though it won’t be released until March 2019, we’re already testing OpenJDK 12 early-access builds against our 18.3 and pre-19.1 builds.)
Since older OpenJDK releases will not receive public support from Oracle (i.e., no security patches), LabKey will stop supporting them shortly after they’re end-of-lifed. To ensure you have all the latest Java security patches and bug fixes, you must regularly upgrade to the latest Java runtime release. You’ll need to upgrade to each six-month feature release plus the intervening security releases (two or three per feature release… roughly every two months).
Based on the published Java release schedule, Java support in LabKey Server for the next year will likely roll-out as follows (where “Java X” means “OpenJDK X and Oracle Java SE X”):
| LabKey Release |
Java Versions Supported |
Changes |
| 18.3 – Nov 2018 |
Java 11, Oracle Java SE 8 |
Add Java 11 |
| 19.1 – Mar 2019 |
Java 11 & 12 |
Add Java 12, Remove Java 8 |
| 19.2 – Jul 2019 |
Java 12 |
Remove Java 11 |
| 19.3 – Nov 2019 |
Java 12 & 13 |
Add Java 13 |
You can always visit our Supported Technologies page to review the latest plan.
Since OpenJDK is a true open-source project, many organizations other than Oracle are now building, distributing, and supporting it. A few of the most prominent examples:
- AdoptOpenJDK has promised community-driven LTS builds of OpenJDK
- Red Hat and other Linux distributions provide and support OpenJDK
- Azul Zulu, IBM, SAP, et al offer free and commercial options
- Amazon recently announced Corretto, a no-cost distribution of OpenJDK that includes long-term support
All of these implementations derive from the same OpenJDK source, so, in theory, they should be interchangeable. However, it’s important to understand that LabKey has not yet tested any of the non-Oracle distributions and, therefore, we do not support them. We plan to test more of these distributions over the next year. But for now, you will need to utilize one of the two Oracle distributions.
Recommendations for Your Team
It’s time to take action on these changes. Java 11 is here and Java 12 is coming soon. Java 8 will reach “End of Public Updates” for commercial users in January 2019, and will stop being a viable option for most deployments. Our recommendations:
- Discuss Java licensing with your legal, licensing, and IT teams. They may have already put in place a runtime subscription or developed a policy around Java. If not, they need to understand these changes and create a plan.
- Based on your organization’s policies, choose the runtime that’s appropriate for your deployments going forward: Oracle OpenJDK 11 or Oracle Java SE 11.
- Upgrade to LabKey 18.3 and then switch to that Java 11 runtime.
- Keep upgrading your runtime… every two months to stay secure.
- If you’re building Java modules, switch your development and testing to JDK 11. IntelliJ makes it easy to configure multiple JDKs and switch between them, if you still need to build with JDK 8 for other work.
Comments? Suggestions? Questions? Please share them on the LabKey support forum .





 Providing support for multi-tabular excel output files and converting them into easy-to-read grids
Providing support for multi-tabular excel output files and converting them into easy-to-read grids LabKey Server’s Luminex data management tools help labs improve quality control of their data by:
LabKey Server’s Luminex data management tools help labs improve quality control of their data by: Logging changes to data records and allowing scientists to view the history of data transformations from the raw file to the analyzed results
Logging changes to data records and allowing scientists to view the history of data transformations from the raw file to the analyzed results

 Integrating Heterogeneous Data
Integrating Heterogeneous Data Creating an Adaptable Database
Creating an Adaptable Database Real-Time Data Sharing
Real-Time Data Sharing