For life science organizations, evaluating and selecting a data management system is a critical process that can have a lasting impact on their research. Research data is among the most valuable assets that an organization possesses due to the enormous amount of time, effort, and resources spent in its collection and analysis. Whether from lab instruments, a clinical trial or an observational study, data is central to decision-making, and ultimately to scientific discovery.
When evaluating data management systems, focus on scalability and flexibility.
Efficiently managing research data requires a data management system that is specifically tailored to the ever-changing needs of scientific research. Planning for the future needs of your organization is essential when choosing a data management system. Research organizations may need to plan for:
- Expanding their lab, research team or collaborators
- Shifting into new experiments, instruments, assays, therapeutic areas or types of data
- Dealing with exponential growth in the volume of data being captured
- Experiencing increased needs for compliance, security, privacy, data sharing, provenance tracking, reproducibility
The data management system you choose should be able to handle any of the scenarios above. By focusing on your future needs you can avoid being locked into a system that will not grow and adapt to new requirements. Under this guiding principle, Our recent webinar outlined six areas that are critical to choosing the right data management system for your organization.
When evaluating a data management system look for:
- Robust security and compliance controls
The first requirement for any data management system is that it meets the security and compliance needs of your organization and area of research. Without meeting these requirements the system will likely hit internal roadblocks and may be deemed insufficient for your organization and use case. - Ability to handle large volumes of diverse data
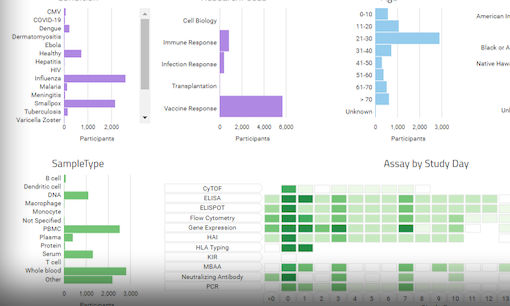
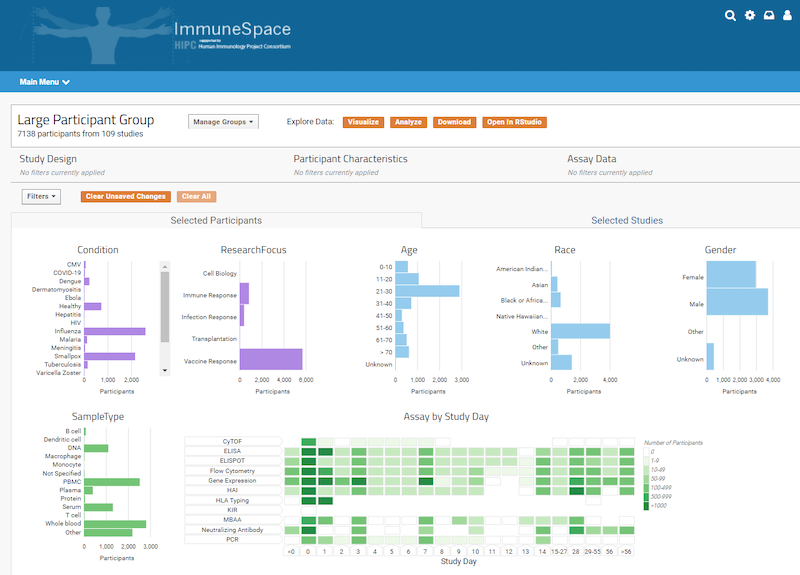
Your data management system should be designed to efficiently bring together and integrate large volumes (millions of rows, hundreds of columns) of diverse data. A clinical trial may include demographics data, sample data, instrument data, and clinical data. Your system should be able to centralize and align all of these data types for efficient and accurate analysis. - Support for automated, high throughput data acquisition
Large volumes of data necessitate automation for scalable and efficient workflows. However, data files are often imperfect and automating their import requires robust quality control functions to ensure the accuracy and integrity of data within the system. - Integration of disparate data and metadata
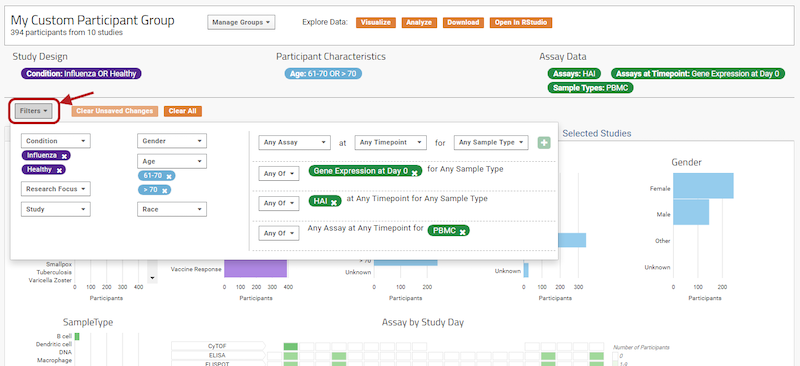
Beyond acquiring large volumes of data using automation, the system must integrate and annotate data in scientifically relevant ways. For example, a clinical trial may need to align sample, assay, and clinical data via participants and visits. By using clinical ontologies, your system should be able to harmonize your data in meaningful ways. - Support for data analytics systems
When deciding what system support is required for analytic tools, it is imperative that you include your research team and the larger organization. The system you choose should support a wide diversity of analytic tools. This includes both native tools within the system as well as third-party analysis software. - Actively developed and supported by the vendor
Choosing a system that is actively being developed and supported by the vendor is key to the ability of the system to grow and adapt to the changing needs of your organization. Like scientific research, technology is consistently evolving and your vendor should be dedicated to the success of your organization and research goals.
Watch the webinar below to learn more:[vc_video link=”https://youtu.be/jIubcWkiPwo” align=”center”]