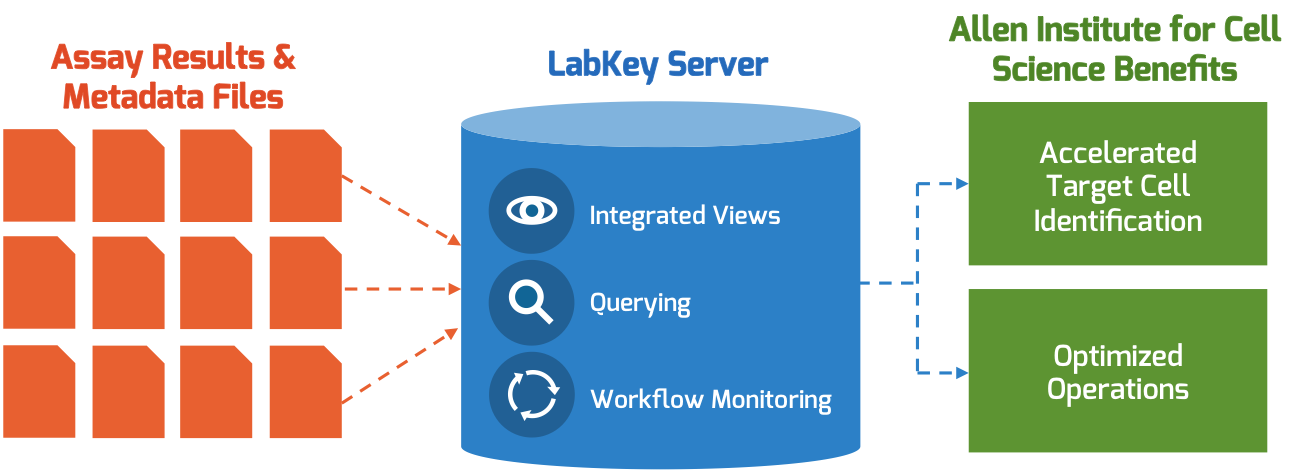
2017 was a year of many milestones for the LabKey team. Our newest product, LabKey Biologics, launched in April and has been implemented at four leading pharma and biotech companies, we redesigned the LabKey Server user interface, and we took our compliance functionality to the next level with the introduction of compliant hosting and a suite of new features to support teams working within HIPAA, FISMA and CFR Part 11 compliant environments. LabKey is excited about what the next year has in-store and we want to share our vision for 2018 with the growing LabKey community.

Reinvesting Premium Dollars into Premium Functionality
Each year, LabKey invests in a number of development projects that premium subscribers have shared as key priorities. In 2018, we plan to dedicate development time to a variety of key areas including:
- Helping secure access to protected data with added support for PHI/PII handling
- Opening the power of R to more users in a secure manner by introducing sandboxing of R code execution
- Increasing the reliability and reproducibility of results via versioning of R engines and packages within RStudio configurations
- Streamlining data integration and enabling rich querying by adding support for clinical ontologies and controlled vocabularies
- Automating and expanding data acquisition options through pipeline file watchers and integration with cloud storage providers
Curious what new functionality was introduced in 2017? Read our “What’s New” updates >
 Delivering on the Biologics Roadmap
Delivering on the Biologics Roadmap
The LabKey Biologics team, in partnership with our product advisory council, has developed an aggressive roadmap for continued development of this flexible system for biologic discovery and development. In 2018, the team will be focusing much of their efforts on:
- Simplifying access to all relevant experiment data by enabling the grouping of samples, assay data, and notes by experiment
- Helping teams better understand their data by expanding reporting and visualization capabilities
- Making it easier for users to standardize and track the creation of media batches through usability improvements to the application’s media registration tools
Interested in learning more about LabKey Biologics? Watch our “Quick Look” video series >
 Strengthening our Core Technology Infrastructure
Strengthening our Core Technology Infrastructure
In order to support our ongoing commitment to providing reliable software solutions, each year we dedicate resources to evolving our technology infrastructure to ensure its on-going stability. This year, we are largely focused on enhancements that will improve efficiency for both internal and external developers working with LabKey Server. Our key priorities include the development of a method for sharing JavaScript components between modules, fully supporting Java 9, and maintaining full compatibility with new versions of third-party components.
 Refining the LabKey Server User Interface, According to User Feedback
Refining the LabKey Server User Interface, According to User Feedback
We have heard from new users of LabKey Server that the current experience makes it difficult to get started using the platform. In order to make sure new users can be successful using LabKey Server from the get go, our UX team will be working with new users over the next few months to understand how we can improve our user interface and the tools we provide. In addition, we will be continuing to expand our our hosted solutions in 2018 to make it easier for organizations to adopt and support LabKey Server.
Want to learn more about the recent LabKey Server UI changes? Check out our “New LabKey Server UI” blog series >
In addition to strengthening our solutions this year, we want to continue to strengthen our relationships with you, the LabKey community. We hope that you will join us in this journey toward creating robust and powerful tools for accelerating research. A few ways that you can get involved this year include:
Providing Feedback on the New UI
As teams upgrade to LabKey Server version 17.3 and later, we encourage you to provide feedback about the new UI through the “Give Feedback” form that can be accessed in your LabKey Server account menu. This feedback is reviewed, prioritized, and used directly to help us make decisions about refinements to the LabKey Server UI.
Participating in a New User Walk-Through
Are you new to using LabKey Server? We want to hear about your experience and how we can improve! LabKey User Experience Manager, Jessi Murray, is looking for new users who would be willing to discuss their experience getting started with LabKey Server and how it could be improved.
Email Jessi Murray at jessim@labkey.com for more information!
Attending the LabKey User Conference
The LabKey User Conference is an opportunity for users to gather together and share about how they are using and extending the LabKey platform and solutions. Attendees also participate in round table discussions and work sessions with the LabKey developers where they can explore current challenges and shared feedback directly with the LabKey team.
Interested in speaking at the LabKey User Conference? Email conference manager, Kelsey Gibson at kelseyg@labkey.com for more information!

 In late 2015,
In late 2015,